Project leaders: Dr Surafel Tegegne and Professor Mark Febbraio
Modern biology generates vast amounts of data — from genes and proteins to metabolites and cellular signals. The challenge isn't collecting this information; it's making sense of it. This project harnesses artificial intelligence and machine learning to integrate multi-omics data, uncovering patterns invisible to traditional analysis and identifying novel disease biomarkers and therapeutic targets.
Understanding disease through data integration
Metabolic diseases like metabolic dysfunction-associated steatohepatitis (MASH), obesity and diabetes don't arise from single causes — they develop through complex interactions between genetics, diet, physical activity and metabolism. By investigating how diet and physical activity influence molecular mechanisms across multiple biological layers (genomics, transcriptomics, proteomics, metabolomics), we're deepening our understanding of these conditions at an unprecedented level of detail.
A data-driven approach to MASH
Our research pipeline integrates experimental data from mouse models with clinical data from human patients, using AI to identify predictive biomarkers and disease mechanisms. This approach moves us toward:
- Non-invasive diagnostics
Identifying blood-based biomarkers that could replace liver biopsies for MASH diagnosis. - Early prediction
Detecting metabolic liver disease before irreversible damage occurs. - Precision medicine
Tailoring treatments based on individual molecular profiles. - Novel therapeutic targets
Discovering new pathways that could be targeted with drugs.
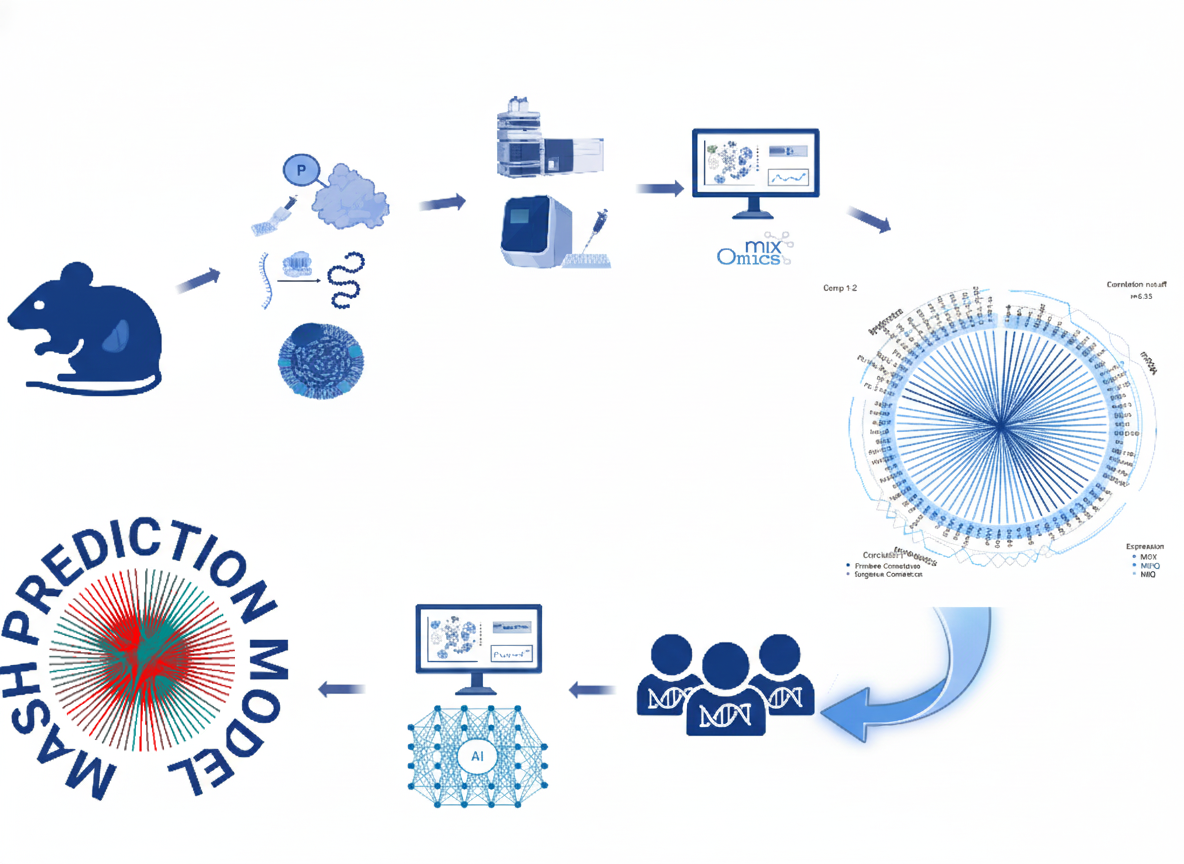
Figure: This study provides a data-driven approach for MASH diagnosis and prediction, integrating experimental (mouse) and clinical (human) data. The workflow shows: mouse models → multi-omics profiling → mixOmics analysis → correlation networks → human cohorts → AI-powered prediction models. AI-powered biomarker discovery could lead to non-invasive diagnostic tools and inform precision medicine strategies for metabolic liver disease.
What makes this approach powerful
Traditional research typically examines one type of biological data at a time — for example, looking at gene expression or protein levels in isolation. Multi-omics integration combines these different data layers simultaneously, revealing how changes at one level (such as genes) cascade through to others (proteins, metabolites, cellular function).
AI and machine learning excel at finding patterns in these complex, high-dimensional datasets — patterns that would be impossible for researchers to identify manually. By training algorithms on both mouse and human data, we can discover biomarkers and mechanisms that translate across species, accelerating the path from laboratory discoveries to clinical applications.
Tools for the community
Beyond our own research, we're developing web-based applications to support extracellular vesicle biology research. These tools will be freely available to the broader scientific community, helping researchers worldwide analyse complex omics data from EV studies and accelerating discoveries in this rapidly evolving field.
Research applications
This AI-driven approach is being applied to understand:
- How exercise and diet modify molecular pathways that protect against metabolic disease.
- Why some individuals develop MASH whilst others with similar risk factors don't.
- Which molecular signatures predict disease progression or treatment response.
- How extracellular vesicles mediate the beneficial effects of lifestyle interventions.
Next steps
Our ongoing work focuses on:
- Validating AI-identified biomarkers in independent clinical cohorts.
- Developing accessible diagnostic tools based on discovered biomarkers.
- Creating publicly available web applications for omics data analysis.
- Expanding the approach to additional metabolic conditions.
By combining cutting-edge AI technology with rigorous biological research, we're transforming how we understand, diagnose and ultimately treat metabolic diseases.